EpiRhandbook の bookdown 実行のスクラップ
-
Ubuntu 22.04 LTS で EpiRhandbook をコンパイルする手順
-
bookdown を使うための準備
-
EpiRHandbook を bookdown でコンパイルするための最低限のパッケージ
-
install_packages.Rとして保存しておく
-
必要な OS パッケージのインストール
$ sudo apt update &&\
sudo apt install -y \
cmake \
build-essential \
libxml2-dev \
libudunits2-dev \
libgdal-dev \
libgeos-dev \
libproj-dev \
libglpk-dev \
libsodium-dev \
gdal-bin \
gdal-data \
pktools-dev \
libmagick++-dev
- OpenstreetMap パッケージのインストールには rJava パッケージのインストールが必要
- rJava パッケージ のインストールには JDK が必要
$ sudo apt install openjdk-18-jdk
$ # openjdk-18 を指定する場合
$ # sudo R CMD javareconf JAVA_HOME=/usr/lib/jvm/java-18-openjdk-amd64/ -e
$ R CMD javareconf -e
# 起動したシェルで R を起動
$ R
> # 起動した R session 内でパッケージのインストールを実行
> source("install_packages.R")
> q() # 一旦セッションを終了する
改めて R セッションを起動し、bookdown を実行する
> bookdown::render_book(input = '.', output_format = 'bookdown::bs4_book', clean = TRUE, config_file = '_bookdown.yml')
指定の章のみのレンダリング
> bookdown::render_book(input = '.', output_format = 'bookdown::bs4_book', clean = TRUE, config_file = '_small_bookdown.yml')
Ubuntu 22.04 LTS で実行するときに、ファイルの大文字小文字、空白の扱いの違いでいくつかのファイル名の変換が必要
$ cp -a ./images/RStudio_overview.PNG ./images/RStudio_overview.png
$ cp -a ./images/markdown/8_ppttemplate.PNG ./images/markdown/8_ppttemplate.png
$ cp -a './images/Epi R Handbook Banner Beige 1500x500.png' ./images/Epi_R_Handbook_Banner_Beige_1500x500.png
$ cp -a './images/Epi interview image.jpg' ./images/Epi_interview_image.jpg
$ cp -a './images/Epi R Handbook Banner Japanese 1500x500.png' ./images/Epi_R_Handbook_Banner_Japanese_1500x500.png
-
index.Rmdファイル内の該当の画像ファイル名を変更しておく
- Docker image を使って環境構築
rocker/rstudio:4.3.2 の docker image を利用
$ docker run -it --rm --mount type=bind,source="$(pwd)",target=/opt/epiRhandbook_jp rocker/rstudio:4.3.2 /bin/bash
# 別シェルを立ち上げて変更をコミット
# docker commit <image id> <repository:tag>
# docker push <repository:tag>
$ docker commit <image id> ysaito8015/epirhandbook_jp:versionTag
$ docker push ysaito8015/epirhandbook_jp:versionTag
$ cat /etc/lsb-release
DISTRIB_ID=Ubuntu
DISTRIB_RELEASE=22.04
DISTRIB_CODENAME=jammy
DISTRIB_DESCRIPTION="Ubuntu 22.04.3 LTS"
$ apt update &&\
apt dist-upgrade -y &&\
apt install -y \
cmake \
build-essential \
pandoc \
libxml2-dev \
libudunits2-dev \
libgdal-dev \
libgeos-dev \
libproj-dev \
libglpk-dev \
libsodium-dev \
gdal-bin \
gdal-data \
pktools-dev \
libmagick++-dev \
openjdk-18-jdk \
libharfbuzz-dev \
libfribidi-dev
$ R CMD javareconf -e
javareconf -e で起動したシェル内で引き続き作業する
# R パッケージのインストールを済ませておく
$ Rscript install_packages.R 2>&1 | tee ./log/install_packages.log
以下のスクリプトを、bookdoen_build.R で保存しておく
bookdown::render_book(
input = '.',
output_format = 'bookdown::bs4_book',
clean = TRUE,
config_file = '_bookdown.yml'
)
log を記録しながら実行
$ Rscript bookdown_build.R 2>&1 | tee ./log/bookdown_build.log
パッケージのインストールでエラーが起きている
Warning messages:
1: In utils::install.packages(pkg_download[!installed_packages]) :
installation of package ‘ClassDiscovery’ had non-zero exit status
2: In utils::install.packages(pkg_download[!installed_packages]) :
installation of package ‘PCDimension’ had non-zero exit status
> pacman::p_load("ClassDiscovery")
Installing package into ‘/usr/local/lib/R/site-library’
(as ‘lib’ is unspecified)
Warning: dependency ‘Biobase’ is not available
trying URL 'https://packagemanager.rstudio.com/cran/__linux__/focal/latest/src/contrib/ClassDiscovery_3.4.0.tar.gz'
Content type 'binary/octet-stream' length 506131 bytes (494 KB)
==================================================
downloaded 494 KB
ERROR: dependency ‘Biobase’ is not available for package ‘ClassDiscovery’
* removing ‘/usr/local/lib/R/site-library/ClassDiscovery’
The downloaded source packages are in
‘/tmp/RtmpVbROUE/downloaded_packages’
Warning messages:
1: In utils::install.packages(package, ...) :
installation of package ‘ClassDiscovery’ had non-zero exit status
Biobase がないと
とりあえずインストール
> BiocManager::install("Biobase", update = TRUE, ask = FALSE)
もしくは
> BiocManager::install("ClassDiscovery", update = TRUE, ask = FALSE, force = TRUE)
> pacman::p_load("PCDimension")
Installing package into ‘/usr/local/lib/R/site-library’
(as ‘lib’ is unspecified)
trying URL 'https://packagemanager.rstudio.com/cran/__linux__/focal/latest/src/contrib/PCDimension_1.1.13.tar.gz'
Content type 'binary/octet-stream' length 197878 bytes (193 KB)
==================================================
downloaded 193 KB
* installing *source* package ‘PCDimension’ ...
** package ‘PCDimension’ successfully unpacked and MD5 sums checked
** using staged installation
** R
** data
** inst
** byte-compile and prepare package for lazy loading
** help
*** installing help indices
** building package indices
** installing vignettes
** testing if installed package can be loaded from temporary location
** testing if installed package can be loaded from final location
** testing if installed package keeps a record of temporary installation path
* DONE (PCDimension)
The downloaded source packages are in
‘/tmp/RtmpVbROUE/downloaded_packages’
PCDimension installed
もう一度 install_packages.R を実行して出てきたエラー
R を再起動で解決
Failed with error: ‘lazy-load database '/usr/local/lib/R/site-library/flextable/R/flextable.rdb' is corrupt’
In addition: Warning messages:
1: In FUN(X[[i]], ...) : restarting interrupted promise evaluation
2: In FUN(X[[i]], ...) : internal error -3 in R_decompress1
Warning message:
In pacman::p_load(bookdown, downlit, xml2, here, dplyr, incidence2, :
Failed to install/load:
downlit
renv について
install
> install.packages(c("renv", "yaml"))
restore
> renv::restore()
document
asdf で R をバージョン管理する
$ asdf plugin-add R https://github.com/asdf-community/asdf-r.git
$ R_EXTRA_CONFIGURE_OPTIONS='--enable-R-shlib --with-cairo' asdf install R 4.2.1
bookdown 実行時のエラー
label: unnamed-chunk-210
Quitting from lines 5182-5189 (_main.Rmd)
Error: 'clean_variable_spelling' is not an exported object from 'namespace:linelist'
In addition: Warning messages:
1: 278 missing rows were removed (86 values from `age_cat5` and 278 values from `gender`).
2: 278 missing rows were removed (86 values from `age_cat5` and 278 values from `gender`).
3: 278 missing rows were removed (86 values from `age_cat5` and 278 values from `gender`).
linelist <- linelist %>%
linelist::clean_variable_spelling(
wordlists = cleaning_dict,
spelling_vars = "col",
# 参照する列名を含む列を指定する
#(デフォルトではディクショナリの3番目の列になる)
)
- エラー発生
- 該当のパッケージをインストールして解決
- Ubuntu 22.04 LTS
- R 4.2.1
- Terminal
label: read_climate (with options)
List of 2
$ warning: logi FALSE
$ message: logi FALSE
Quitting from lines 16075-16086 (_main.Rmd)
Error in read_ncdf(file_paths[1]) :
package ncmeta required, please install it first
In addition: There were 26 warnings (use warnings() to see them)
-
エラー発生
- 上のエラーを解決して、パッケージインストール用スクリプトを実行した後
-
Ubuntu 22.04 LTS
-
R 4.2.1
-
Terminal
label: unnamed-chunk-697 (with options)
List of 5
$ echo : logi FALSE
$ fig.show : chr "hold"
$ message : logi FALSE
$ warning : logi FALSE
$ out.width: chr [1:2] "50%" "50%"
Quitting from lines 12579-12670 (_main.Rmd)
Error in bold(., i = 6, bold = TRUE, part = "body") :
unused arguments (i = 6, bold = TRUE, part = "body")
5848 In addition: There were 22 warnings (use warnings() to see them)
参考
- http://124.219.182.167/epiR_Build_20220719.html
- http://124.219.182.167/epiR_BuildIssues_20220727.html
重要
-
_bookdown_filesをコンパイル前に必ず削除する
-
bookdown::render_book()にclean = TRUEオブションを渡すと毎回削除してくれる
> bookdown::render_book(
input = '.',
output_format = 'bookdown::bs4_book',
clean = TRUE,
config_file = '_bookdown.yml'
)
-
エラー発生
-
環境 1
- Ubuntu 22.04 LTS
- R 4.2.1
- Terminal
-
環境 2
- Windows 10 Pro
- R 4.2.1
- RStudio RStudio-2022.07.1-554.exe
label: read_climate (with options)
List of 2
$ warning: logi FALSE
$ message: logi FALSE
no 'var' specified, using t2m, tp
other available variables:
longitude, latitude, time
Will return stars object with 8760 cells.
No projection information found in nc file.
Coordinate variable units found to be degrees,
assuming WGS84 Lat/Lon.
no 'var' specified, using t2m, tp
other available variables:
longitude, latitude, time
Will return stars object with 8760 cells.
No projection information found in nc file.
Coordinate variable units found to be degrees,
assuming WGS84 Lat/Lon.
Quitting from lines 16075-16086 (_main.Rmd)
Error in Ops.stars(read_ncdf(file_paths[1]), read_ncdf(file_paths[2])) :
(first) dimensions of e1 and e2 do not match
In addition: There were 26 warnings (use warnings() to see them)```該当チャンクの実行結果
> read_ncdf(file_paths[1])
no 'var' specified, using t2m, tp
other available variables:
longitude, latitude, time
Will return stars object with 8760 cells.
No projection information found in nc file.
Coordinate variable units found to be degrees,
assuming WGS84 Lat/Lon.
stars object with 3 dimensions and 2 attributes
attribute(s):
Min. 1st Qu. Median Mean 3rd Qu. Max.
t2m [K] 256.459 276.7418 2.816750e+02 2.818962e+02 2.874753e+02 3.049785e+02
tp [m] 0.000 0.0000 2.328388e-06 1.137138e-04 5.704550e-05 8.477014e-03
dimension(s):
from to offset delta refsys point values x/y
longitude 1 1 NA NA WGS 84 NA 10.423 [x]
latitude 1 1 NA NA WGS 84 NA 51.334 [y]
time 1 8760 2002-01-01 UTC 1 hours POSIXct NA NULL
> read_ncdf(file_paths[2])
no 'var' specified, using t2m, tp
other available variables:
longitude, latitude, time
Will return stars object with 8760 cells.
No projection information found in nc file.
Coordinate variable units found to be degrees,
assuming WGS84 Lat/Lon.
stars object with 3 dimensions and 2 attributes
attribute(s):
Min. 1st Qu. Median Mean 3rd Qu. Max.
t2m [K] 258.7856 275.4088 2.813136e+02 2.819685e+02 2.883852e+02 3.076843e+02
tp [m] 0.0000 0.0000 1.273918e-07 6.300391e-05 2.181585e-05 4.174184e-03
dimension(s):
from to offset delta refsys point values x/y
longitude 1 1 NA NA WGS 84 NA 10.423 [x]
latitude 1 1 NA NA WGS 84 NA 51.334 [y]
time 1 8760 2003-01-01 UTC 1 hours POSIXct NA NULL
> (read_ncdf(file_paths[1]) + read_ncdf(file_paths[2]))
no 'var' specified, using t2m, tp
other available variables:
longitude, latitude, time
Will return stars object with 8760 cells.
No projection information found in nc file.
Coordinate variable units found to be degrees,
assuming WGS84 Lat/Lon.
no 'var' specified, using t2m, tp
other available variables:
longitude, latitude, time
Will return stars object with 8760 cells.
No projection information found in nc file.
Coordinate variable units found to be degrees,
assuming WGS84 Lat/Lon.
Error in Ops.stars(read_ncdf(file_paths[1]), read_ncdf(file_paths[2])) :
(first) dimensions of e1 and e2 do not match
次元はあってるっぽいのに
> a <- stars::read_ncdf(file_paths[1])
no 'var' specified, using t2m, tp
other available variables:
longitude, latitude, time
Will return stars object with 8760 cells.
No projection information found in nc file.
Coordinate variable units found to be degrees,
assuming WGS84 Lat/Lon.
> b <- stars::read_ncdf(file_paths[2])
no 'var' specified, using t2m, tp
other available variables:
longitude, latitude, time
Will return stars object with 8760 cells.
No projection information found in nc file.
Coordinate variable units found to be degrees,
assuming WGS84 Lat/Lon.
> dim(a)
longitude latitude time
1 1 8760
> dim(b)
longitude latitude time
1 1 8760
該当行を下記にすると、実行可能
data <- stars::read_stars(file_paths)
実際に bookdown を実行して、公開する環境に合わせたほうが良さそう
- エラーの発生
- 環境
- Windows 10 Pro
- R 4.2.1
- RStudio RStudio-2022.07.1-554.exe
Quitting from lines 9888-9895 (_main.Rmd)
Error in makePSOCKcluster(names = spec, ...) :
numeric 'names' must be >= 1
In addition: Warning messages:
1: 278 missing rows were removed (86 values from `age_cat5` and 278 values from `gender`).
2: 278 missing rows were removed (86 values from `age_cat5` and 278 values from `gender`).
3: 278 missing rows were removed (86 values from `age_cat5` and 278 values from `gender`).
4: Removed 256 rows containing non-finite values (stat_bin).
5: Removed 686 rows containing missing values (position_stack).
6: Removed 2744 rows containing missing values (position_stack).
7: Removed 2058 rows containing missing values (position_stack).
====================
fastLink(): Fast Probabilistic Record Linkage
====================
If you set return.all to FALSE, you will not be able to calculate a confusion table as a summary statistic.
Calculating matches for each variable.
Error in makePSOCKcluster(names = spec, ...) :
numeric 'names' must be >= 1
-
エラー発生
-
環境 1
- Ubuntu 22.04 LTS
- R 4.2.1
- Terminal
-
環境 2
label: observation_time
Quitting from lines 19166-19210 (_main.Rmd)
Error in find_end_date(., "left_date", "death_date", period_start = "recall_start", :
could not find function "find_end_date"
In addition: There were 50 or more warnings (use warnings() to see the first 50)
epikit パッケージからエクスポートされている関数がみつからない、と
該当の関数にパッケージ名を付加した
> sitrep::find_end_date()
> sitrep::find_start_date()
次のエラー発生
label: observation_time
Quitting from lines 19167-19211 (_main.Rmd)
Error: object 'add_weights_cluster' is not exported by 'namespace:epikit'
In addition: There were 50 or more warnings (use warnings() to see the first 50)
knitr::knit2html() はできるのに、bookdown ではできない。。。なぜ。。。
> knitr::knit2html(input="./new_pages/survival_analysis.Rmd")
Could not find function "..." について
library() で読み込ませてみるか...
-
クリーンインストールしてビルドを試した Windows 環境でも同じエラー発生
-
環境 3
- Windows 10 Pro 20H2
- R 4.2.1
- RStudio 2022.07.01-554
label: observation_time
Quitting from lines 19166-19210 (_main.Rmd)
Error in find_end_date(., "left_date", "death_date", period_start = "recall_start", :
could not find function "find_end_date"
In addition: There were 50 or more warnings (use warnings() to see the first 50)
sessioninfo::session::info() で比較中
windows 環境と rocker/rstudio 環境のエラーは、sitrep パッケージをインストールすることで解決
> install.packages("drat", repos = "https://cran.rstudio.com")
> drat::addRepo("R4EPI")
> install.packages("sitrep")
-
エラー発生
-
環境
label: read_climate (with options)
List of 2
$ warning: logi FALSE
$ message: logi FALSE
Quitting from lines 16059-16070 (_main.Rmd)
Error in dyn.load(file, DLLpath = DLLpath, ...) :
unable to load shared object '/usr/local/lib/R/site-library/units/libs/units.so':
libudunits2.so.0: cannot open shared object file: No such file or directory
apt 実行
$ apt update && \
apt install -y libudunits2-dev libgdal-dev libgeos-dev libproj-dev
-
エラー発生
-
環境
label: unnamed-chunk-855 (with options)
List of 7
$ out.width : chr [1:2] "100%" "100%"
$ fig.show : chr "hold"
$ echo : logi FALSE
$ fig.width : num 12
$ fig.height: num 9
$ message : logi FALSE
$ warning : logi FALSE
Quitting from lines 17397-17483 (_main.Rmd)
Error in make_epicontacts(linelist = linelist, contacts = contacts, directed = TRUE) :
could not find function "make_epicontacts"
In addition: There were 50 or more warnings (use warnings() to see the first 50)
> epicontacts::make_epicontacts()
Error in dyn.load(file, DLLpath = DLLpath, ...) :
unable to load shared object '/usr/local/lib/R/site-library/igraph/libs/igraph.so':
libglpk.so.40: cannot open shared object file: No such file or directory
該当のシステムライブラリをインストール
$ apt update && apt install -y libglpk-dev
-
エラーの発生
-
環境
- Windows 10 Pro
- R 4.2.1
- RStudio RStudio-2022.07.1-554.exe
label: unnamed-chunk-980 (with options)
List of 2
$ message: logi FALSE
$ warning: logi FALSE
Quitting from lines 21434-21443 (_main.Rmd)
Error in openmap(upperLeft = c(max(linelist$lat, na.rm = T), max(linelist$lon, :
could not find function "openmap"
Registered S3 method overwritten by 'quantmod':
method from
as.zoo.data.frame zoo
Registered S3 method overwritten by 'ape':
method from
plot.mst spdepIn addition: There were 50 or more warnings (use warnings() to see the first 50)
rJava パッケージが入っていなかった
> # 緯度経度の範囲に基図を切り取る。タイル種別を選択。
> map <- OpenStreetMap::openmap(
+ upperLeft = c(max(linelist$lat, na.rm=T), max(linelist$lon, na.rm=T)), # 基図のタイルの制限
+ lowerRight = c(min(linelist$lat, na.rm=T), min(linelist$lon, na.rm=T)),
+ zoom = NULL,
+ type = c("osm", "stamen-toner", "stamen-terrain", "stamen-watercolor", "esri","esri-topo")[1])
Error: .onLoad は loadNamespace()('rJava' に対する)の中で失敗しました、詳細は:
call: fun(libname, pkgname)
error: JAVA_HOME cannot be determined from the Registry
-
エラー発生
-
環境
label: epidemic_models_incubation_estimate_epiestim (with options)
List of 1
$ warning: logi FALSE
Quitting from lines 17825-17827 (_main.Rmd)
Error in fit_disc_gamma(get_pairwise(epic, "date_onset")) :
lazy-load database '/usr/local/lib/R/site-library/epicontacts/R/epicontacts.rdb' is corrupt
In addition: There were 50 or more warnings (use warnings() to see the first 50)
-
エラーの発生
-
環境
- Windows 10 Pro
- R 4.2.1
- RStudio RStudio-2022.07.1-554.exe
label: unnamed-chunk-1361 (with options)
List of 3
$ out.width: chr [1:2] "25%" "25%"
$ fig.show : chr "hold"
$ echo : logi FALSE
Quitting from lines 29530-29589 (_main.Rmd)
Error in pal(nlev) : could not find function "pal"
In addition: There were 50 or more warnings (use warnings() to see the first 50)
-
エラー発生
-
環境1
- Ubuntu 22.04 LTS
- R 4.2.1
- Terminal
-
環境2
-
環境3
- Windows 10 Pro
- R 4.2.1
- RStudio RStudio-2022.07.1-554.exe
label: unnamed-chunk-944
Quitting from lines 20962-20966 (_main.Rmd)
Error in find_formula.data.frame(x, verbose = FALSE) :
A data frame is not a valid object for this function.
In addition: There were 50 or more warnings (use warnings() to see the first 50)
insight パッケージの find_formula.data.flame()
このエラーで詰まってしまう
ん?エラーが微妙に変わった
label: unnamed-chunk-943
Quitting from lines 20951-20955 (_main.Rmd)
Error: A data frame is not a valid object for this function.
In addition: There were 50 or more warnings (use warnings() to see the first 50)
Registered S3 method overwritten by 'ape':
method from
plot.mst spdep
unnamed-chunk-943 に変わっている場合は, missing_data.Rmd の章タイトルと最初の R chunk がなぜか省かれている
message() を使ってデバグ
Ubuntu, Docker 環境は、_bookdown.yml から gis.Rmd をコメントアウトすると、それ以外のファイルは、問題なくビルドされる
Windows 環境は、なぜか missing_data.Rmd の冒頭10行ほどが _main.Rmd に出力されない
該当のチャンクで使われている janitor::clean_names() のドキュメント https://www.rdocumentation.org/packages/janitor/versions/1.2.0/topics/clean_names
以下で実行
dplyr::filter(sle_adm3_raw, "admin2Name" %in% c("Western Area Urban", "Western Area Rural"))
エラー
label: clean_adm3 (with options)
List of 1
$ message: logi FALSE
Quitting from lines 20966-20971 (_main.Rmd)
Error in UseMethod("filter") :
no applicable method for 'filter' applied to an object of class "logical"
In addition: There were 50 or more warnings (use warnings() to see the first 50)
rstudio でのスレッド https://community.rstudio.com/t/filter-doesnt-run-no-applicable-method-for-filter-applied-to-an-object-of-class-logical/98305
あー。わかったかも。
janitor::clean_names() とするとそのチャンクが通るということは
パッケージがアタッチされていない?
Windows で EpiRHandbook をビルドする手順まとめ
-
rJava パッケージのインストールに必要な JDK をインストールする
- windows 用インストーラを選択してダウンロード https://www.oracle.com/java/technologies/downloads/#jdk18-windows
- EpiRHandbook を bookdown でコンパイルするための最低限のパッケージを R Console に入力
- R セッションを再起動する
- 次のコマンドを入力
> bookdown::render_book(input = '.', output_format = 'bookdown::bs4_book', clean = TRUE, config_file = '_bookdown.yml')
マシンの性能にもよりますが、2−3時間はかかるかもしれません
bookdown::render_book() したときのワーニング
[WARNING] Duplicate identifier 'flexdashboard' at _main.knit.md line 44539 column 1
章タグが new_pages/data_used.Rmd のセクションタグと重複していたので、該当部分を変更
PR https://github.com/appliedepi/epiRhandbook_jp/pull/73
Windows 10 の環境でビルドした html_output に missing_data.Rmd の出力が含まれていない問題発生
_main.Rmd 内に misisng_data.Rmd の章タイトルと数行が含まれていない
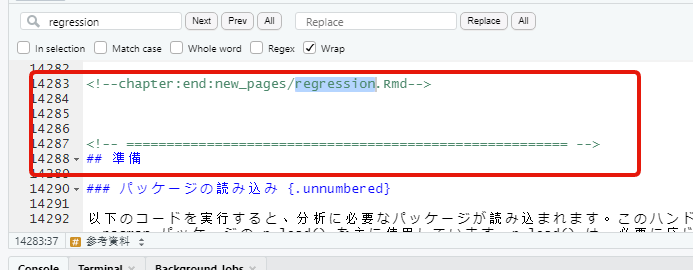
Docker image でビルドした方には入っている。。。
わからない。。。
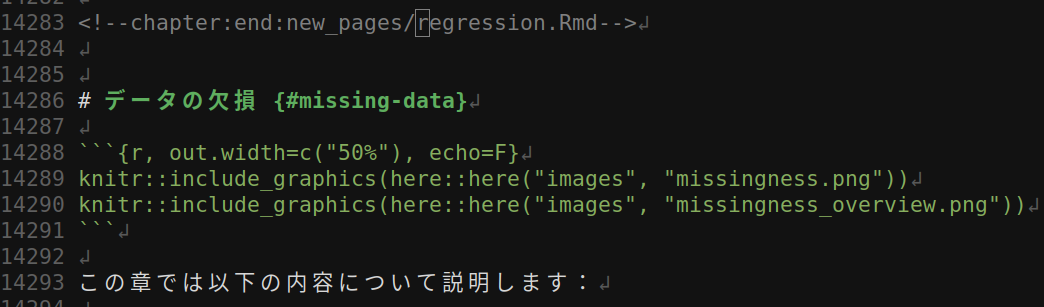
windows 環境でビルドする場合のみ発生する、固有の問題のようなので
docker image でビルドすることで回避
英語版 https://github.com/appliedepi/epiRhandbook_eng/ での windows 環境の bookdown ビルドでは、正しく missing_data.Rmd の冒頭数行が入っているので、
日本語版固有の問題っぽい
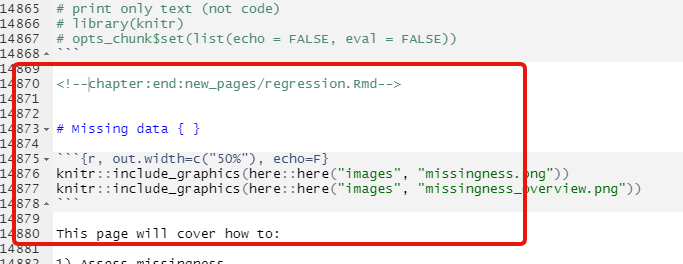
エラー発生
- 環境3
- Windows 10 Pro
- R 4.2.1
- RStudio RStudio-2022.07.1-554.exe
label: epidemic_models_epiestim_config
Quitting from lines 17813-17818 (_main.Rmd)
Error in make_config(mean_si = 12, std_si = 5.2) :
could not find function "make_config"
In addition: There were 50 or more warnings (use warnings() to see the first 50)
エラー
label: visualise_population_flow
Quitting from lines 19612-19650 (_main.Rmd)
Error in `mutate()`:
! Problem while computing `x = fct_relevel(x, c("startcause", "endcause"))`.
Caused by error in `fct_relevel()`:
! `.f` must be a factor or character vector, not an integer vector
Backtrace:
1. ... %>% ...
7. forcats::fct_relevel(x, c("startcause", "endcause"))
英語版 v1.0.1 へ追従する更新をして、bookdown 実行
エラー
- 環境2
label: unnamed-chunk-856 (with options)
List of 7
$ out.width : chr [1:2] "100%" "100%"
$ fig.show : chr "hold"
$ echo : logi FALSE
$ fig.width : num 12
$ fig.height: num 9
$ message : logi FALSE
$ warning : logi FALSE
Quitting from lines 17474-17560 (_main.Rmd)
Error in xy.coords(x, y, xlabel, ylabel, log) :
'x' is a list, but does not have components 'x' and 'y'
Calls: <Anonymous> ... eval -> eval -> plot -> plot -> plot.default -> xy.coords
Execution halted
該当箇所
エピデミックモデリング
## パッケージのインストールとロード
pacman::p_load(tidyverse, EpiNow2, EpiEstim, here, incidence2, epicontacts, rio, projections)
## ラインリストのロード
linelist <- import(here::here("data", "case_linelists", "linelist_cleaned.rds"))
## コンタクトの作成
contacts <- linelist %>%
transmute(
from = infector,
to = case_id
) %>%
drop_na()
## エピコンタクトの作成
epic <- make_epicontacts(
linelist = linelist,
contacts = contacts,
directed = TRUE
)
## ## estimate gamma generation time
## generation_time <- bootstrapped_dist_fit(
## get_pairwise(epic, "date_infection"),
## dist = "gamma",
## max_value = 20,
## bootstraps = 1
## )
## ## export for caching
## export(
## generation_time,
## here("data/cache/epidemic_models/generation_time.rds")
## )
## import cached generation time
generation_time <- import(here("data/cache/epidemic_models/generation_time.rds"))
## ## estimate incubation period
## incubation_period <- bootstrapped_dist_fit(
## linelist$date_onset - linelist$date_infection,
## dist = "lognormal",
## max_value = 100,
## bootstraps = 1
## )
## ## export for caching
## export(
## incubation_period,
## here("data/cache/epidemic_models/incubation_period.rds")
## )
## import cached incubation period
incubation_period <- import(here("data/cache/epidemic_models/incubation_period.rds"))
## get incidence from onset date
cases <- linelist %>%
group_by(date = date_onset) %>%
summarise(confirm = n())
## ## run epinow
## epinow_res <- epinow(
## reported_cases = cases,
## generation_time = generation_time,
## delays = delay_opts(incubation_period),
## target_folder = here("data/cache/epidemic_models"),
## return_output = TRUE,
## output = "samples",
## verbose = TRUE,
## stan = stan_opts(samples = 750, chains = 4),
## horizon = 21
## )
## ## export for caching
## export(
## epinow_res,
## here("data/cache/epidemic_models/epinow_res.rds")
## )
## import cached epinow results
epinow_res <- import(here("data/cache/epidemic_models/epinow_res.rds"))
## plot summary figure
plot(epinow_res)
- 該当箇所は英語版リポジトリと差分なし
- x, y のデータの長さが違うとは?
R v4.2.2 ではエラーが出る
R v4.2.1 ではエラーが出ない
英語版 v1.0.1 へ追従する更新をして、bookdown 実行
エラー
- 環境2
label: unnamed-chunk-1303 (with options)
List of 3
$ out.width: chr [1:2] "25%" "25%"
$ fig.show : chr "hold"
$ echo : logi FALSE
Quitting from lines 29609-29671 (_main.Rmd)
Error in epicontacts::thin(., "contacts") :
x is not an epicontacts object
Calls: <Anonymous> ... eval_with_user_handlers -> eval -> eval -> %>% -> <Anonymous>
In addition: There were 50 or more warnings (use warnings() to see the first 50)
Execution halted
- epicontacts パッケージを再インストール
> remotes::install_github("reconhub/epicontacts@timeline")
- igraph パッケージが non-zero exit status
Error: package or namespace load failed for ‘ igraph’ in dyn.load(file, DLLpath = DLLpath, ...):
unable to load shared object '/home/ysaito/.asdf/installs/R/4.2.1/lib/R/library/00LOCK-igraph/00new/igraph/libs/igraph.so':
libgfortran.so.4: cannot open shared object file: No such file or directory
Error: loading failed
Execution halted
ERROR: loading failed
* removing ‘ /home/ysaito/.asdf/installs/R/4.2.1/lib/R/library/igraph’
* restoring previous ‘ /home/ysaito/.asdf/installs/R/4.2.1/lib/R/library/igraph’
The downloaded source packages are in
‘ /tmp/RtmpYOE39D/downloaded_packages’
Updating HTML index of packages in '.Library'
Making 'packages.html' ... done
Warning message:
In install.packages("igraph") :
installation of package ‘ igraph’ had non-zero exit status
$ apt install gfortran-7
エラー解決
うーん
パッケージをアップデートしたあとに同じところで引っかかる
もう一度インストール
> update.packages(ask = FALSE)
> remotes::install_github("reconhub/epicontacts@timeline", force = TRUE)
わかった! epic オブジェクトを subset 化するコードにミスがあった
epic_small <- epic %>%
subset(
node_attribute = list(date_onset = c(as.Date(c("2014-06-01", "2014-07-01"))))) %>%
epicontacts::thin("contacts")
英語版 v1.0.1 へ追従する更新をして、bookdown 実行
エラー
- 環境2
label: visualise_population_flow
Quitting from lines 19634-19658 (_main.Rmd)
Error in `geom_parallel_sets()`:
! Problem while converting geom to grob.
ℹ Error occurred in the 1st layer.
Caused by error in `check.length()`:
! 'gpar' element 'lwd' must not be length 0
Backtrace:
1. bookdown::render_book(...)
2. bookdown:::render_cur_session(...)
3. rmarkdown::render(main, output_format, ..., clean = clean, envir = envir)
4. knitr::knit(knit_input, knit_output, envir = envir, quiet = quiet)
5. knitr:::process_file(text, output)
...
50. grid::gpar(...)
51. grid:::validGP(list(...))
52. grid (local) numnotnull("lwd")
53. grid (local) check.length(gparname)
54. base::stop(...)
Execution halted
特定のパッケージのバージョンの組み合わせで発生するエラーの様子
パッケージをアップデートしたら解決
参考 Error: ! Problem while converting geom to grob. ℹ Error occurred in the 1st layer
英語版 v1.0.1 へ追従する更新をして、bookdown 実行
エラー
- 環境2
label: phylogenetic_trees_adding_sampledata (with options)
List of 3
$ fig.align: chr "center"
$ warning : logi FALSE
$ message : logi FALSE
Quitting from lines 30475-30514 (_main.Rmd)
Error in `geom_segment2()`:
! Problem while converting geom to grob.
ℹ Error occurred in the 4th layer.
Caused by error in `check.length()`:
! 'gpar' element 'lwd' must not be length 0
Backtrace:
1. bookdown::render_book(...)
2. bookdown:::render_cur_session(...)
3. rmarkdown::render(main, output_format, ..., clean = clean, envir = envir)
4. knitr::knit(knit_input, knit_output, envir = envir, quiet = quiet)
5. knitr:::process_file(text, output)
...
49. grid::gpar(...)
50. grid:::validGP(list(...))
51. grid (local) numnotnull("lwd")
52. grid (local) check.length(gparname)
53. base::stop(...)
Execution halted
BiocManager で管理されているパッケージをアップデート
> BiocManager::install(update = TRUE, ask = FALSE)
なにやらエラー
'getOption("repos")' replaces Bioconductor standard repositories, see
'?repositories' for details
replacement repositories:
CRAN: https://packagemanager.rstudio.com/cran/__linux__/focal/latest
Bioconductor version 3.15 (BiocManager 1.30.19), R 4.2.1 (2022-06-23)
Old packages: 'ggtree'
Update all/some/none? [a/s/n]: a
trying URL 'https://bioconductor.org/packages/3.15/bioc/src/contrib/ggtree_3.4.4.tar.gz'
Content type 'application/x-gzip' length 356386 bytes (348 KB)
==================================================
downloaded 348 KB
* installing *source* package ‘ggtree’ ...
** using staged installation
** R
** inst
** byte-compile and prepare package for lazy loading
Error in get(x, envir = ns, inherits = FALSE) :
object 'warning_wrap' not found
Error: unable to load R code in package ‘ggtree’
Execution halted
ERROR: lazy loading failed for package ‘ggtree’
* removing ‘/usr/local/lib/R/site-library/ggtree’
* restoring previous ‘/usr/local/lib/R/site-library/ggtree’
The downloaded source packages are in
‘/tmp/RtmpHOCJiG/downloaded_packages’
Warning message:
In install.packages(update[instlib == l, "Package"], l, contriburl = contriburl, :
installation of package ‘ggtree’ had non-zero exit status
github からのインストールに変更
> remotes::install_github('YuLab-SMU/ggtree')
エラー解決
ここまでのエラー対応で html がビルドできた
R 4.3.2 に対応するために作業中
- Docker rocker:rstudio:4.3.2 でエラー
Quitting from lines 11228-11251 [unnamed-chunk-599] (_main.Rmd)
Error:
! object 'gender' not found
Backtrace:
1. incidence2::incidence(...)
Execution halted
-
iteration.Rmdの L249-254 -
enderを"gender"へ
outbreak <- incidence2::incidence(
x = linelist, # データフレームを指定する(linelist全体)
date_index = "date_onset", # 日付の列
interval = "week", # 週ごとに集計
groups = "gender") # 性別によるグルーピング
#na_as_group = TRUE) # 性別の欠損値は欠損のグループ(NA)として扱う
- Docker rocker:rstudio:4.3.2 でエラー
Quitting from lines 30055-30058 [unnamed-chunk-1301] (_main.Rmd)
Error in `tree_subset()`:
! could not find function "tree_subset"
Execution halted
l
- 同様のエラー
Quitting from lines 30071-30075 [unnamed-chunk-1303] (_main.Rmd)
Error in `tree_subset()`:
! could not find function "tree_subset"
Execution halted
- パッケージ名を追加して解決
sub_tree1 <- tidytree::tree_subset(
tree,
node = 528)
