Python Implementation of Bayesian Linear Regression Class
Introduction
I noticed that there are not many implementations of Bayesian linear regression with correct implementations, and there are few implementations that handle multi-dimensional inputs, so I implemented a user-friendly class. The description basically follows the PRML.
Implemented Class
To be honest, it's not a big implementation, it's only about 50 lines of code...
I'll write the entire class here. The features it has are as follows.
It's really simple.
- Calculate the posterior distribution given the combination of
\phi t - Perform random sampling of the mean and covariance parameters of Bayesian linear regression
- Output the mean and variance of the predictive distribution given
\phi
import numpy as np
from scipy.stats import multivariate_normal
class BeyesLinearRegression:
def __init__(self, mu, S, beta):
self.mu = mu
self.S = S
self.beta = beta
def calc_posterior(self, phi, t):
S_inv = np.linalg.inv(self.S)
if len(phi.shape) == 1:
phi = phi.reshape(1, -1)
t = t.reshape(1, 1)
self.S = np.linalg.inv(S_inv + self.beta * phi.T @ phi)
self.mu = self.S @ (S_inv @ self.mu + np.squeeze(self.beta * phi.T @ t))
def sampling_params(self, n=1, random_state=0):
np.random.seed(random_state)
return np.random.multivariate_normal(self.mu, self.S, n)
def probability(self, x):
dist = multivariate_normal(mean=self.mu, cov=self.S)
return dist.logpdf(x)
def predict(self, phi):
if len(phi.shape) == 1:
phi = phi.reshape(1, -1)
pred = np.array([self.mu.T @ _phi for _phi in phi])
S_pred = np.array([(1 / self.beta) + _phi.T @ self.S @ _phi for _phi in phi])
# Above is a simple implementation.
# This may be better if you want speed.
# pred = self.mu @ phi.T
# S_pred = (1 / self.beta) + np.diag(phi @ self.S @ phi.T)
return pred, S_pred
The entire code is also available on GitHub. (It's only about 50 lines)
About Bayesian Linear Regression
I won't go into the details of the formula derivation, as the following references are more detailed on this.
-
Regression using Bayesian Estimation(Japanese)
Calculation of Posterior Distribution
The important thing is that the distribution is updated as follows. In simple terms,
Predictive Distribution
The predictive distribution is represented as follows. I'll skip the details here too, but the key point is that the distribution for a new point
Using the Implemented Class
From here, let's try Bayesian linear regression using the class I implemented.
The class I created requires you to provide the
So, just input the original data
Regression on Sine Wave Data
Let's start with some toy data.
Data Generation and Feature Function Design
The input data is the true distribution of the sine wave with noise added as the observation data. As for the design of the feature function, it is a composite wave of several frequencies of trigonometric functions. The method x_to_phi vectorizes 10 waves, and _phi represents the composite wave. The amplitudes are the parameters to be estimated by Bayesian linear regression. Mathematically, it's like the image below. (Come to think of it, the first term is zero... it's not needed...)
def x_to_phi(x):
if len(x.shape) == 1:
x = x.reshape(-1, 1)
return np.concatenate([np.sin(2 * np.pi * x * m) for m in range(0, 10)], axis=1)
def _phi(x, params):
return np.array([p * np.sin(2 * np.pi * x * i) for i, p in enumerate(params)]).sum(axis=0)
Here is the actual data generation part.
x = np.arange(0, 1, 0.01)
phi = x_to_phi(x)
e = np.random.randn(len(x)) / 10
y_true = np.sin(2 * np.pi * x)
y = y_true + e
Case of Observing Only One Point
First, let's consider the case where only the 50th data point is observed.
train_idx = 50
x_train = x[train_idx]
phi_train = phi[train_idx]
y_train = y[train_idx]
plt.scatter(x_train, y_train, c='crimson', marker='o', label='observation')
plt.plot(x, y_true, label='true')

Let's go ahead and calculate the posterior distribution for this data.
If you just want to learn, it's just one line like this.
# Initial values for Bayesian linear regression
input_dim = phi.shape[1]
mu = np.zeros(input_dim)
S = np.identity(input_dim)
sigma = 0.1
beta = 1.0 / (sigma ** 2)
# Model definition and training
beyes_linear_model = BeyesLR.BeyesLinearRegression(mu, S, beta)
beyes_linear_model.calc_posterior(phi_train, y_train)
I'll display the wave forms sampled randomly from the posterior distribution in green dotted lines, and the predictive distribution in light blue.
Since we only learned from one point, most of it is shown in blue.

Case of Observing 50 Points
Let's do exactly the same thing, but this time with 50 observation data points. The only difference in the code is that we randomly select 50 indices for train_idx.
The resulting predictive distribution is shown in the figure below.

Regression on Advertising Dataset
Next, let's solve a real-world problem, a multi-dimensional linear regression this time.
Since increasing the dimensionality by feature extraction would make it too hard to understand, I'll make
Input Data
This is the famous Advertising dataset from ISLR.
This time, I'll use the TV and Radio advertising costs as input, and the sales as the response.
Since
def x_to_phi(x, typ='linear', degree=3):
if len(x.shape) == 1:
x = x.reshape(-1, 1)
return np.concatenate([np.ones(x.shape[0]).reshape(-1, 1), x], axis=1)
df = pd.read_csv(ADVERTISING_DATASET)
x = df[['TV', 'Radio']].values
y = df['Sales'].values
phi = x_to_phi(x)
x_train, x_test, phi_train, phi_test, y_train, y_test = \
train_test_split(x, phi, y, train_size=0.05, random_state=0)
All that's left is to train it the same way as before.
input_dim = phi.shape[1]
mu = np.zeros(input_dim)
S = np.identity(input_dim)
sigma = 10
beta = 1.0 / (sigma ** 2)
beyes_linear_model = BeyesLR.BeyesLinearRegression(mu, S, beta)
beyes_linear_model.calc_posterior(phi_train, y_train)
In the code, I set train_size to 0.05, but the regression planes drawn by changing this are shown below. I learned the Bayesian linear regression and extracted 5 planes by random sampling. When the number of training samples is small, nonsensical planes are drawn, but as the data points increase, it converges.
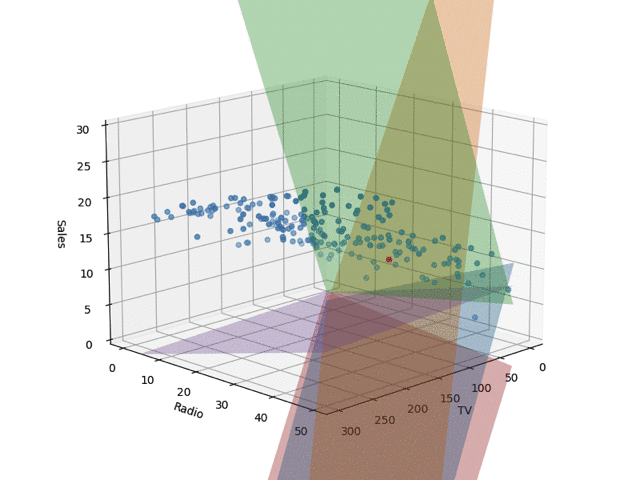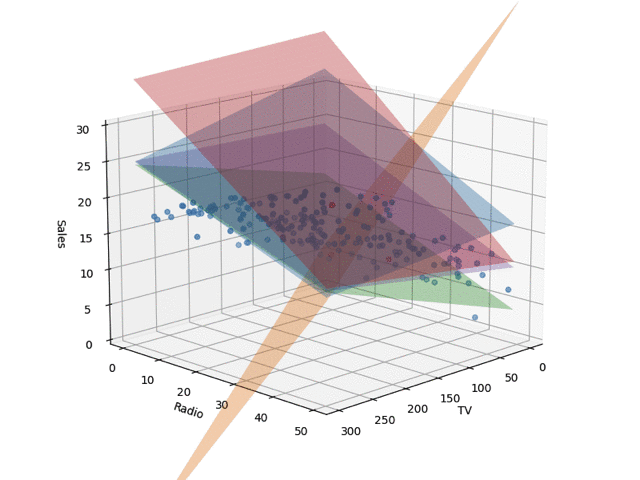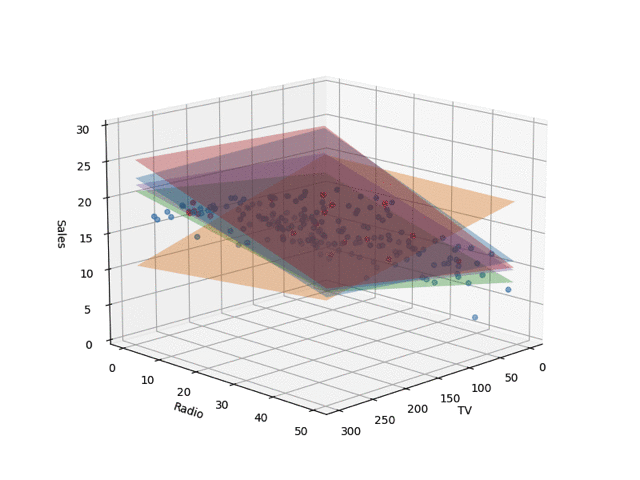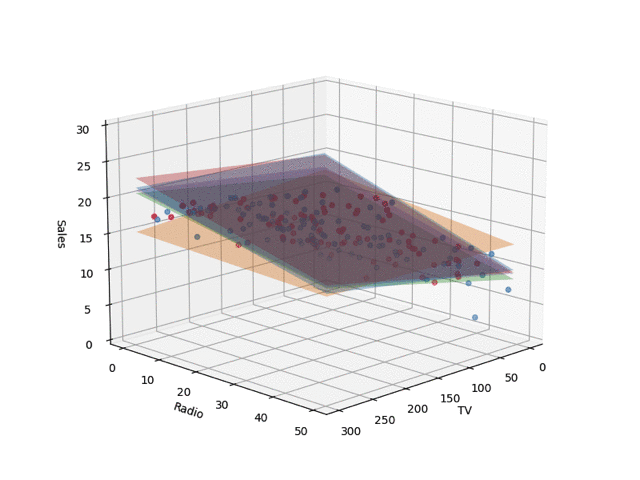
Conclusion
Finally, let me just briefly advertise Bayesian linear regression.
There are still parts that I don't fully understand, but the design of the feature extraction function
However, if you have good prior knowledge to extract good features, Bayesian linear regression can often be sufficient, even from the perspective of interpretability. Also, Bayesian linear regression can do online learning right away due to its calculation method. You just need to use the posterior distribution you calculated as the prior distribution for the next learning. You don't even need to recalculate the Gram matrix like in Gaussian process regression. There are online versions, but...
Well then, I hope you enjoy a comfortable Bayesian linear regression life!

Discussion