How to instal and use smina @M1 Mac mini
When it comes to OSS docking programs, Vina is the classic choice, but its installation process and the preparation of pdbqt files can be quite cumbersome.
Smina and Gnina, forks of Vina, allow for simpler docking studies using pdb or sdf files, making them highly recommended for academia and beginners.
This guide explains how to install Smina and use it on macOS. Apologies for not including Windows instructions 🙇.
Installing Smina
The macOS binary for Smina can be downloaded here. Clicking the link should start the download immediately. Move the file to a convenient location and set up the path for easy terminal-based docking execution. For UNIX-based OS users like macOS or Linux, follow the steps below:
mkdir ~/Documents/apps
mkdir ~/Documents/apps/smina
mv ~/Downloads/smina.osx ~/Documents/apps/smina
cd ~/Documents/apps/smina
mv smina.osx smina
ln -s ~/Documents/apps/smina /usr/local/bin/.
If you're unsure about setting up paths, this guide can be helpful.
Check if the path is properly set up by using the which command:
which smina
# /usr/local/bin/smina
Run smina alone in the terminal, and if the help content for Smina is displayed, the command is successfully ready for use.
Basic Usage of Smina
Smina is a command-line interface (CLI) application. You can refer to this documentation for execution details. Here's a sample command:
smina -r receptor.pdb -l ligand.pdb --autobox_ligand ligand.pdb -o docked.sdf --num_modes 1
Next, let's try docking using a known ligand co-crystal structure with the sample code above.
For testing, we'll use the 6O0P co-crystal structure. Background research on the receptor and ligand is recommended, but for this example, we’re focusing on experimenting with a large ligand.
Run the following commands to prepare the receptor and ligand files:
mkdir ~/Documents/pdb/6o0p # Create a directory where calculation outputs will be saved.
cd ~/Documents/pdb/6o0p
wget https://files.rcsb.org/download/6O0P.pdb
grep ATOM 6O0P.pdb > rec.pdb
grep HETATM 6O0P.pdb > lig.pdb
ls # 6O0P.pdb lig.pdb rec.pdb
This completes the setup. Use ls to confirm the creation of rec.pdb and lig.pdb. You can also visualize the generated files using PyMOL:
pymol lig.pdb rec.pdb
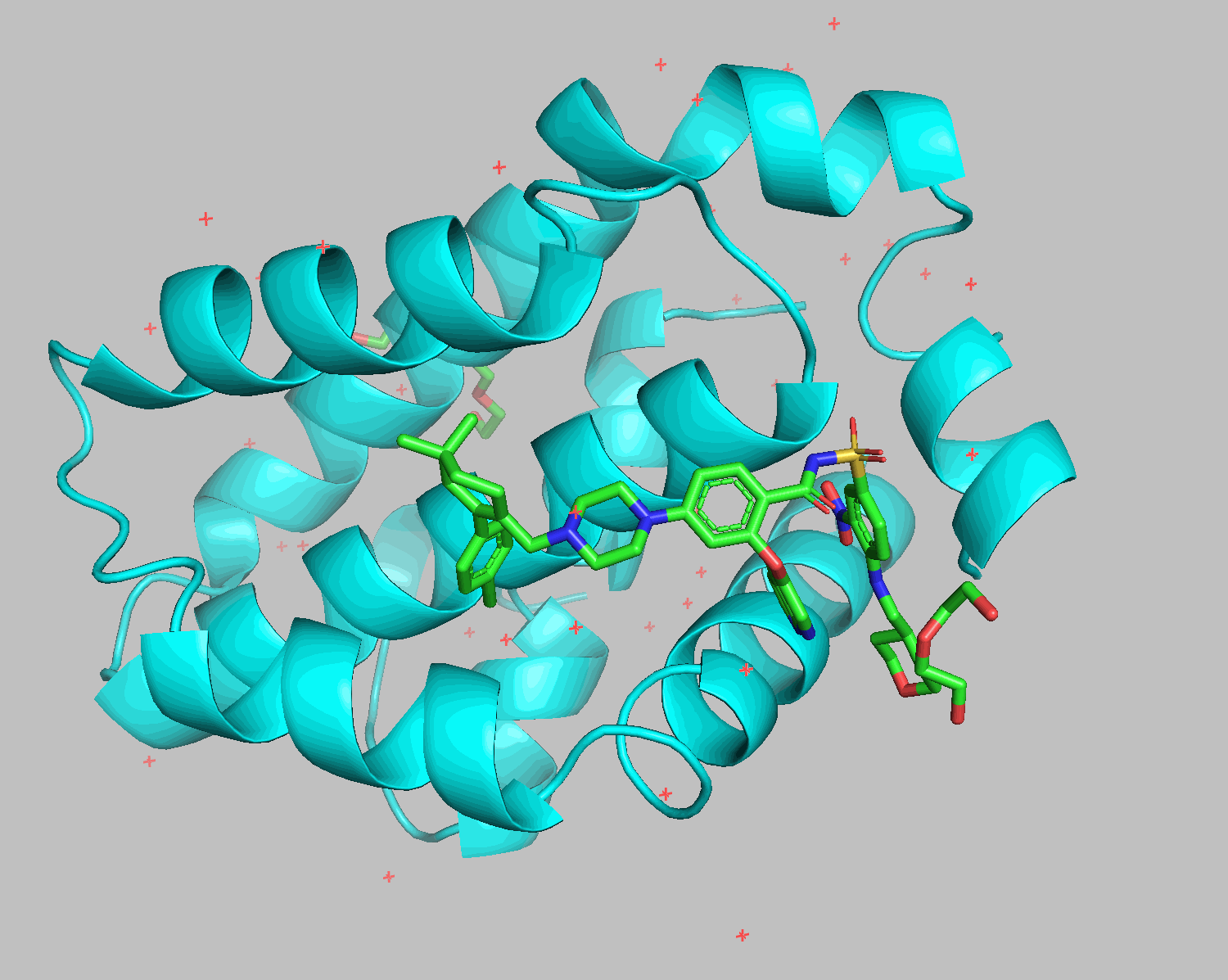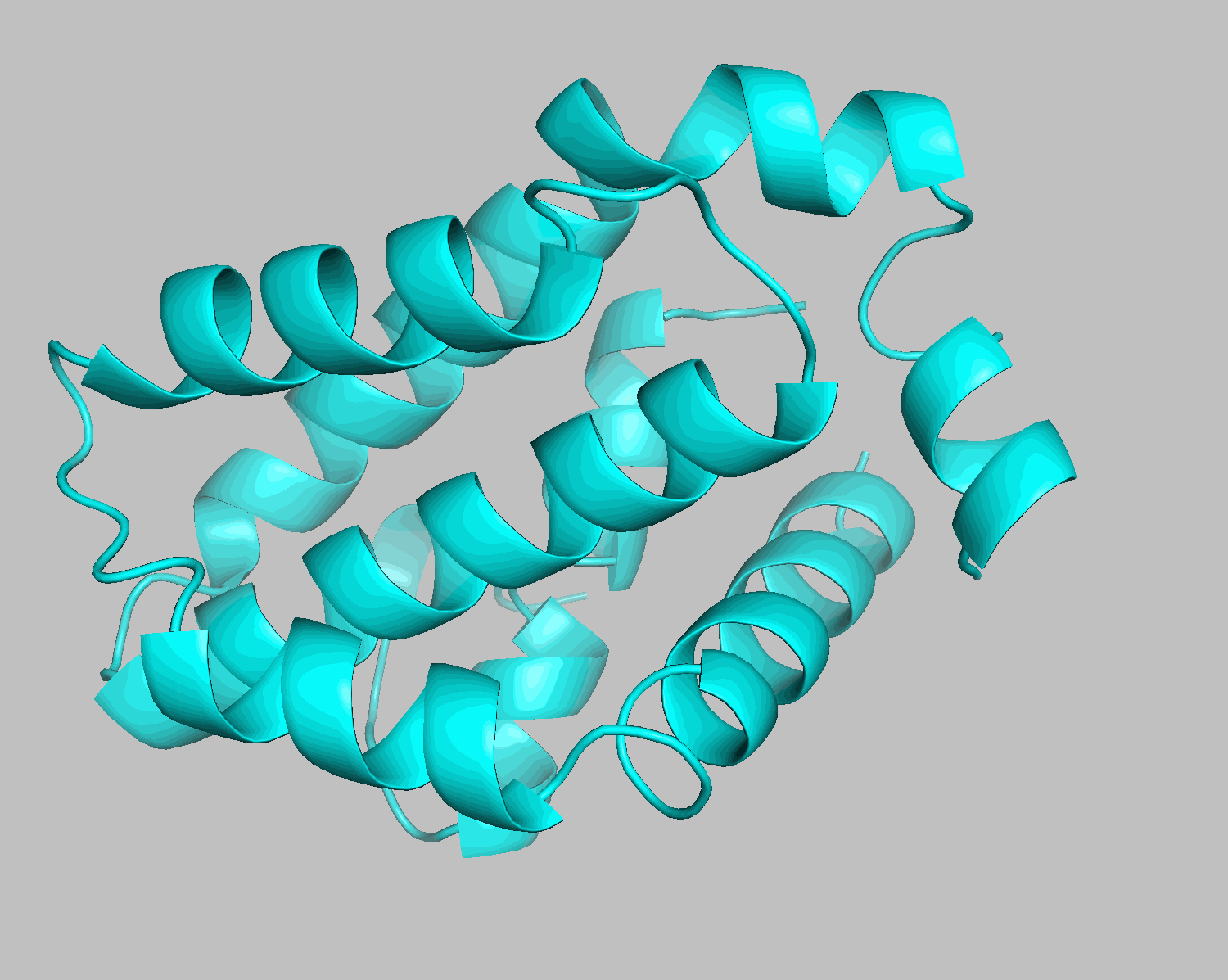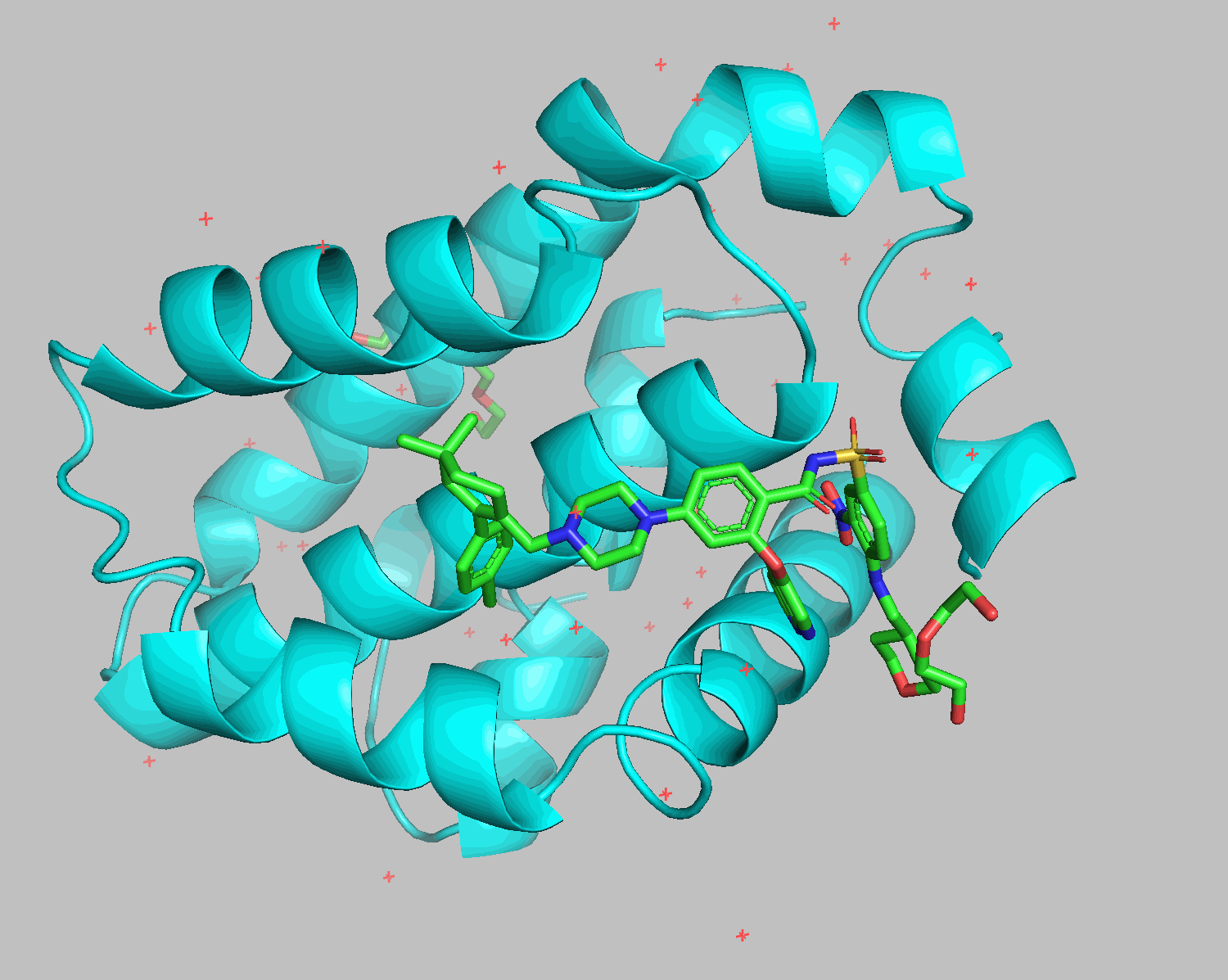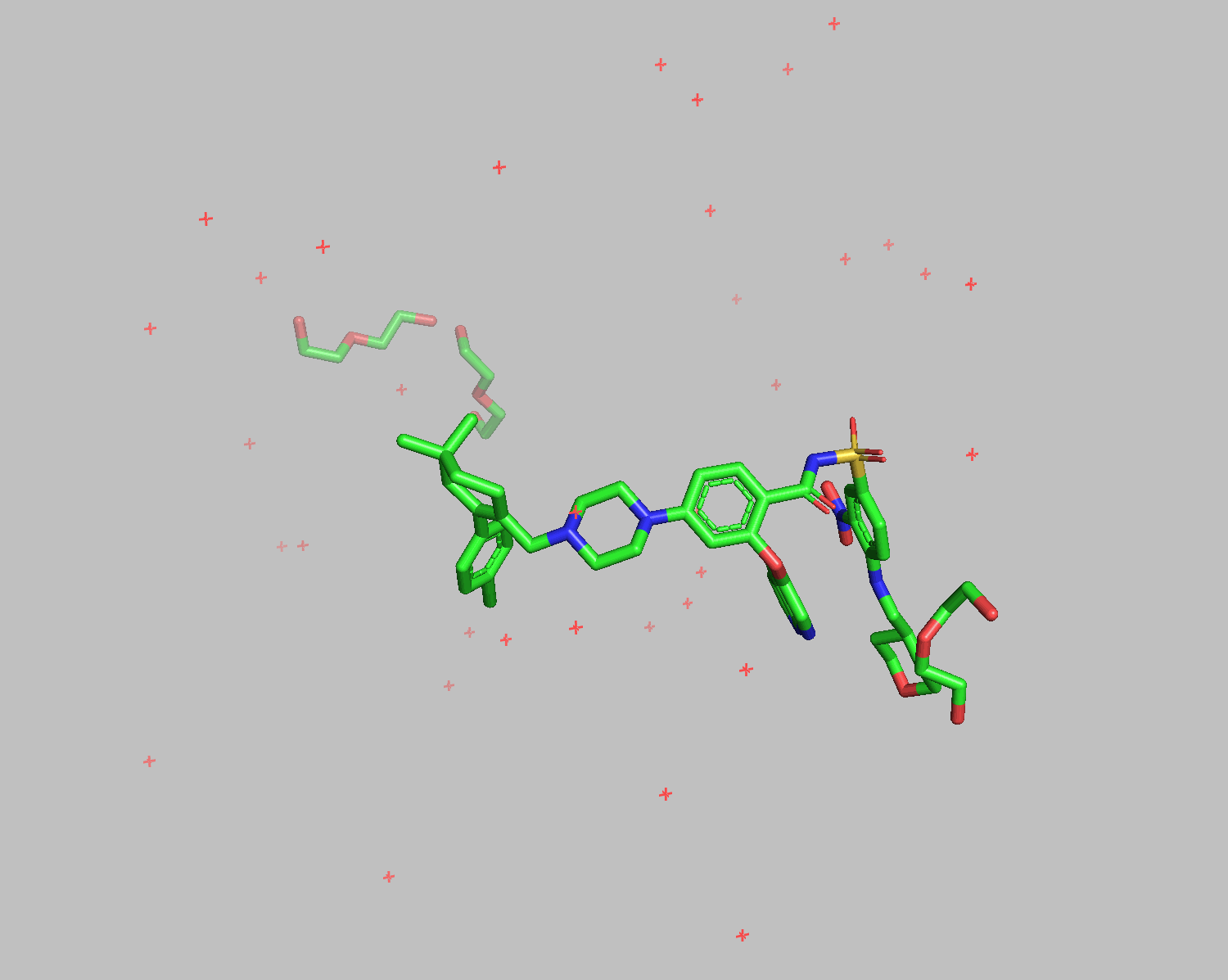
The grep HETATM filename command extracts rows containing HETATM from the specified filename. HETATM in 6O0P.pdb includes not only ligands but also water and additives. One of Smina and Gnina's advantages is their ability to exclude these and dock only the ligand.
Running Smina
Execute Smina by referring to its documentation:
smina -r rec.pdb -l lig.pdb --autobox_ligand lig.pdb -o docked.sdf --num_modes 1 --seed 0
Verify the results using PyMOL. Load the output in PyMOL's command line:
load dock.sdf
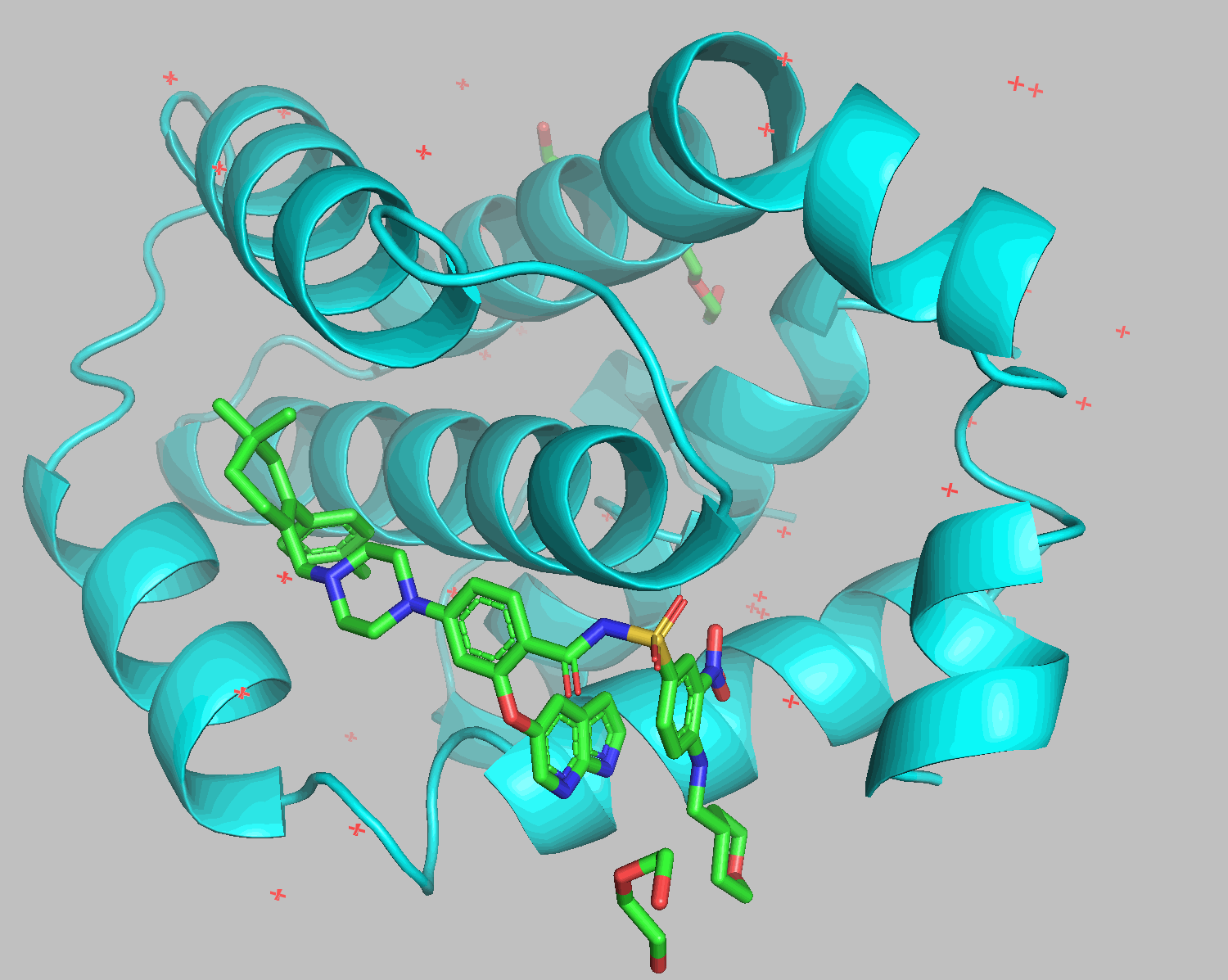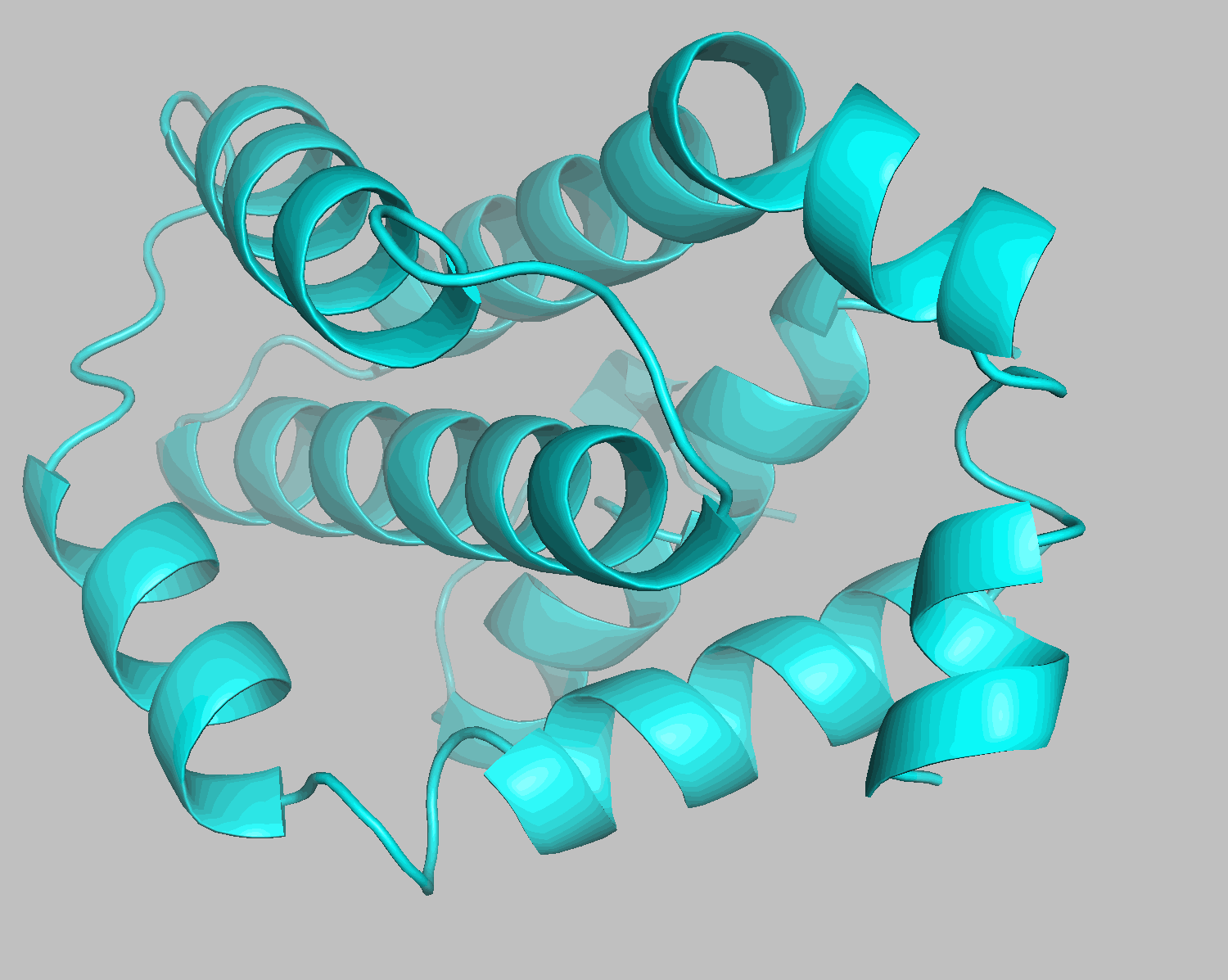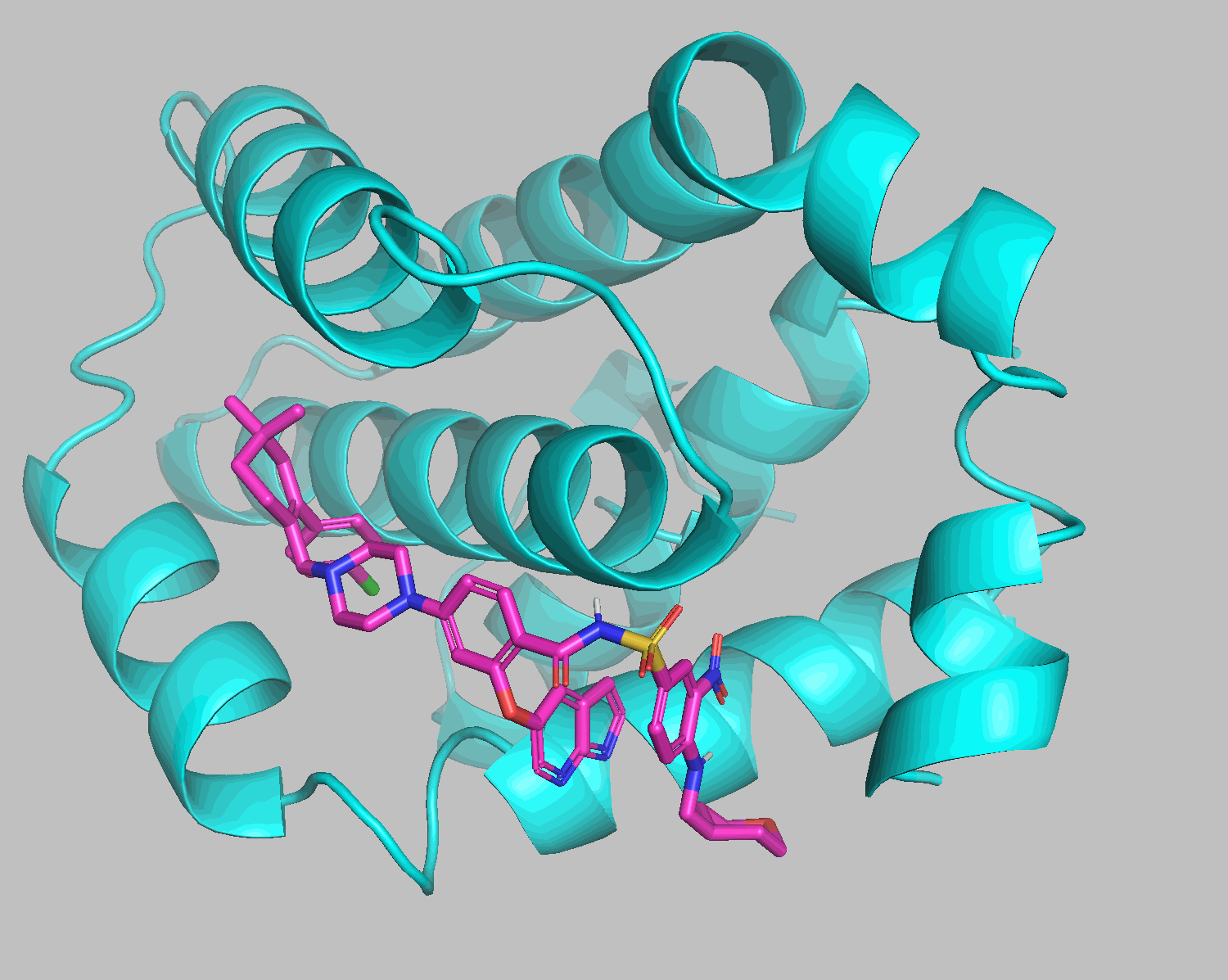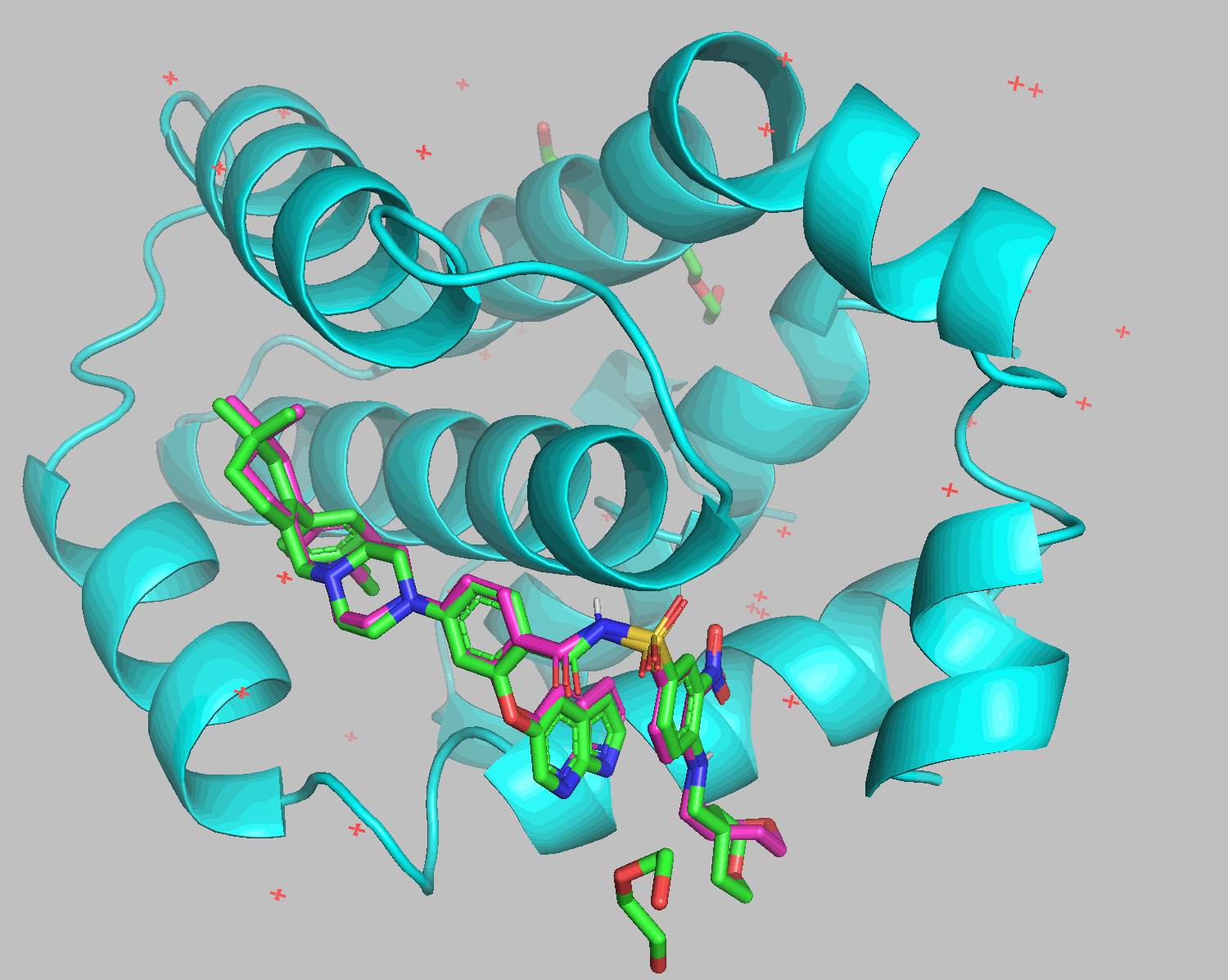
Docking results show that diethylene glycol and water molecules positioned in front of the ligand were excluded. Apart from the THP section on the bottom right, the structures align well.
By using rec.pdb for the --autobox_ligand option, you can explore the entire receptor molecule. For 6O0P, similar results were obtained. Using the same value for the --seed option ensures identical calculation results.
While Vina requires pdbqt files for docking studies, Smina and Gnina streamline the process by supporting pdb files, making them extremely convenient tools.
This article was translated by ChatGPT 4o from my own original contents.
Discussion